Introduction to ToxAI
Toxicity prediction is a vital component of drug discovery, as it directly influences the safety, success, and regulatory approval of potential therapeutic compounds. Assessing toxicity early helps identify harmful side effects such as organ damage, mutagenicity, or cardiotoxicity, ensuring that only safe candidates move forward in the development pipeline.
Late-stage failures due to unforeseen toxic effects are costly and time-consuming, making early in silico screening essential for minimizing risk. To deal with these issues, we have designed an in-house tool, ToxAI. It is an advanced computational tool integrated within the SilicoXplore platform, designed to predict the toxicity of chemical compounds based on their molecular structure.
The tool enables researchers, chemists, and toxicologists to rapidly screen compounds for potential toxicity risks before proceeding with experimental testing, saving valuable time and resources in drug discovery and chemical safety assessment workflows. ToxAI accepts chemical structures in SMILES format and provides comprehensive toxicity predictions and multiple physicochemical property calculations to support decision-making in compound selection and optimization. ToxAI employs a sophisticated Multilayer Perceptron (MLP) Deep Neural Network (DNN) algorithm for toxicity prediction.
A Step-by-Step Guide to Execute the Tool
Before executing the tool, you must create a Job ID. You can customize this ID or click the "Create Job ID" button to have one generated for you automatically.
TIP: Without creating a JOB ID, you will not be able to access any options of the tool.
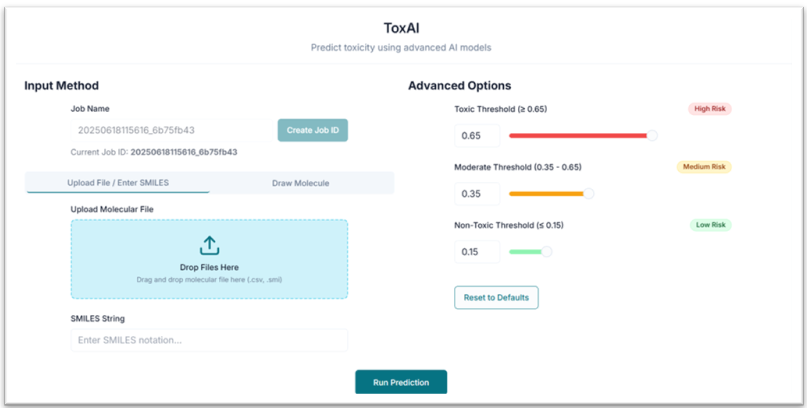
This is the ToxAI tool's application workspace page, where various options for exploring and predicting the toxicity of compounds are displayed. This tutorial will explain each option in detail.
Predicting Toxicity: Providing Input
From the "Upload Your File" tab, you will upload a .smi or .csv file by clicking on the "Drop Files Here" option. The CSV file should contain "Compound name" and "SMILES" columns.
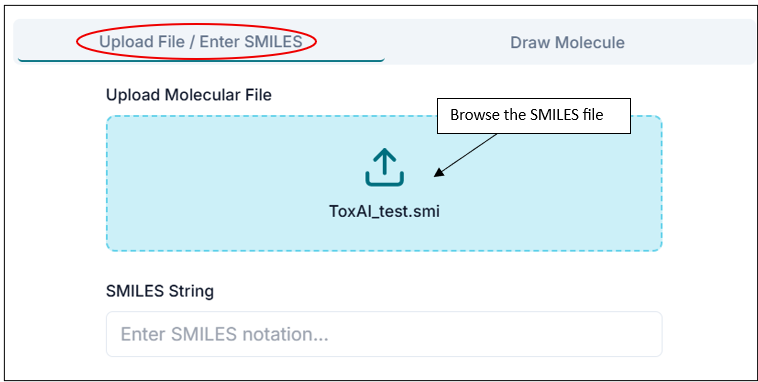
Your input .smi file should be in a similar format to that shown below; it should contain your compound SMILES and their respective names/IDs.

Tip: In .smi files, the SMILES string and the compound name should be TAB-separated.
There is also another option to provide input using the "SMILES Input" tab, where you can paste your SMILES directly.
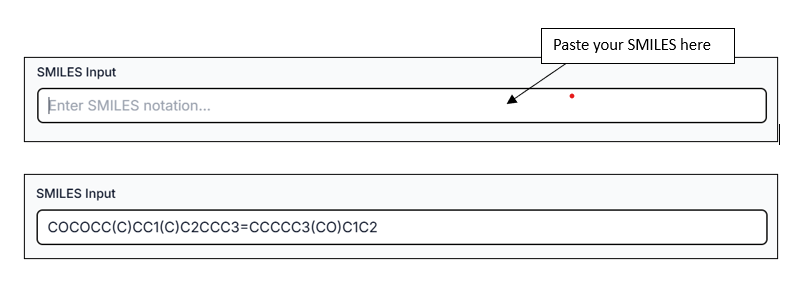
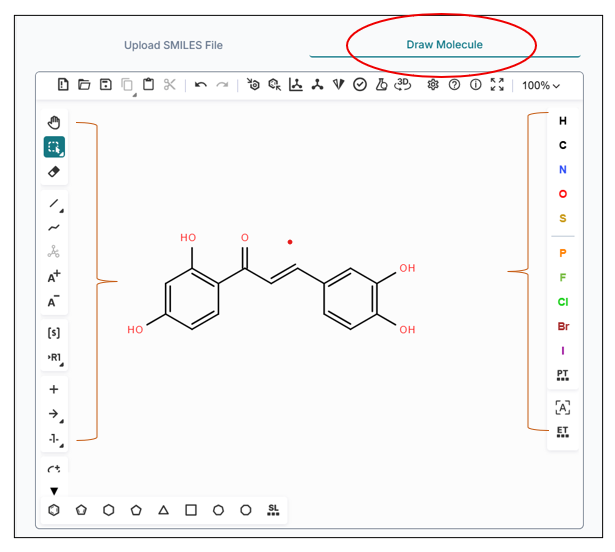
You can also draw any compound using the "Draw Molecule" option from the workspace window. The Draw Molecule workspace gives you the ability to sketch your molecule as you want.
Configuring Thresholds and Running Prediction
After uploading the SMILES file, pasting SMILES, or drawing your compound, you can directly click "Run Prediction". However, if you want to tailor your results, you can first customize the toxicity classification thresholds:
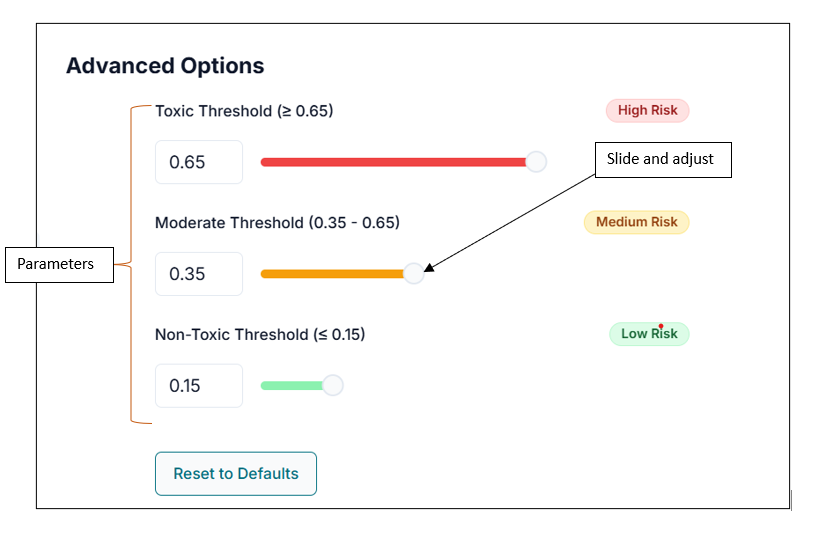
- Toxic Threshold: Set the value above which compounds are classified as toxic (default > 0.65).
- Moderate Threshold: Set the range for compounds classified as moderately toxic (default ≥ 0.35 and < 0.65).
- Non-Toxic Threshold: Set the value below which compounds are classified as non-toxic (default < 0.35).
To submit, click on the "Run Prediction" button.
Analyzing and Downloading Results
After the task is completed, you will be notified by a pop-up as shown below.

You will find the predicted results just below in the application window itself. It shows a summary of how many of the input molecules are classified as Toxic, Non-toxic, and Moderately Toxic.
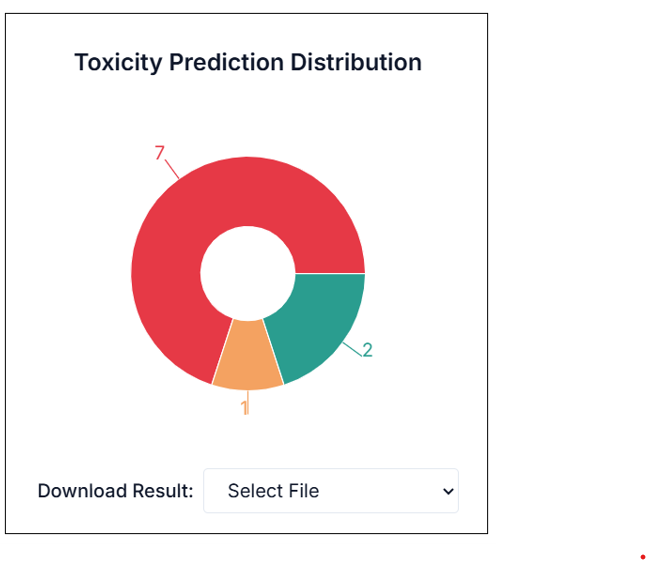
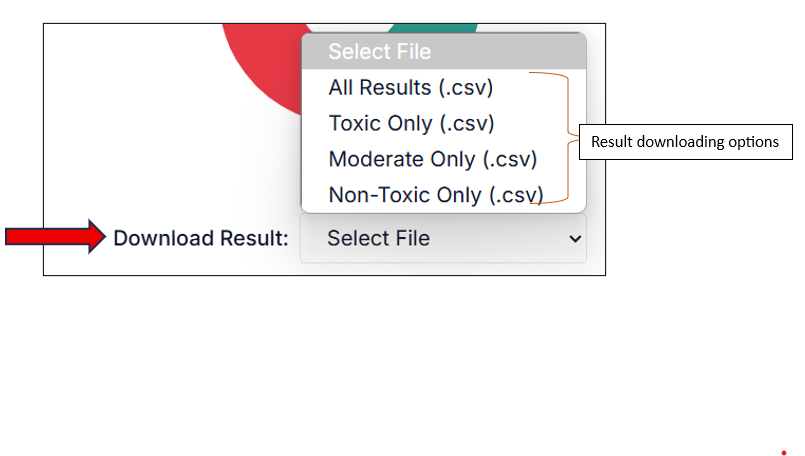
You can download the results from the "Download Result" option. From the drop-down, you can choose which result set you want to download (e.g., all results, only toxic, etc.). The result will be downloaded in a CSV file.
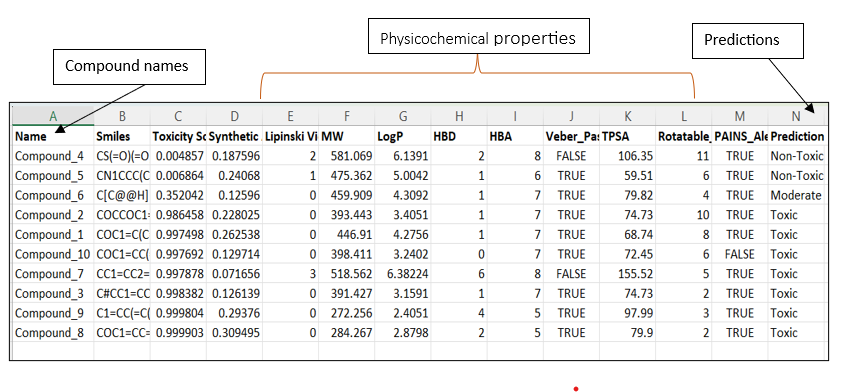
The result CSV file will be as shown in the image below, with all the parameters and the prediction result.