Introduction to Ligand-Interaction
Protein–ligand interactions are central to the mechanism of action for most drugs. A ligand (typically a small molecule or drug) binds to a target protein to modulate its biological activity, either by activating it or inhibiting it. Understanding these interactions is critical for elucidating a drug's mechanism of action, optimizing binding affinity and selectivity, minimizing off-target effects, and designing molecules with improved pharmacological properties.
These interactions form the basis of structure-based drug design (SBDD) and are essential for validating computational predictions from docking and molecular dynamics simulations. To facilitate this, we have designed a tool named Ligand-Interaction. It automatically analyzes 3D protein–ligand complexes (from PDB files or IDs) and identifies key non-covalent interactions such as hydrogen bonds, hydrophobic contacts, π–π stacking, salt bridges, and more. It provides detailed interaction tables, downloadable visualizations, and structured output formats, making it a powerful and user-friendly tool for researchers engaged in molecular modeling and drug design.
A Step-by-Step Guide to Execute the Tool
Before executing the tool, you must create a Job ID. You can customize this ID or click the "Create Job ID" button to have one generated for you automatically.
TIP: Without creating a JOB ID, you will not be able to access any options of the tool.

This is the Ligand-Interaction tool's application workspace page, where various options for exploring and fetching protein-ligand interactions are displayed. This tutorial will explain each option in detail.
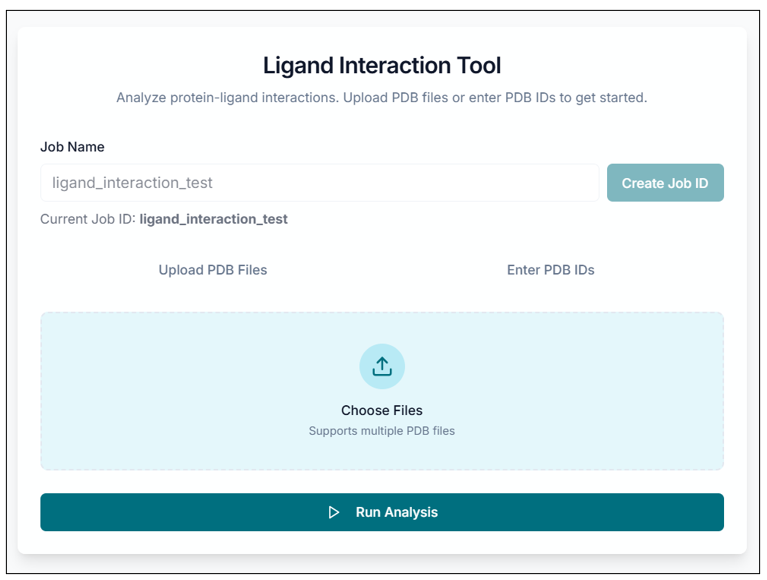
Analyzing a Protein-Ligand Complex
You can upload your protein-ligand complex (.pdb) file for analysis using the "Upload PDB Files" option, or you can directly enter the PDB ID using the "Enter PDB ID" option. The selected file name will be displayed below the upload area.
Tip: You can upload multiple PDB protein-ligand complex files using the "Choose file" option.
After choosing your file, click on the "Run Analysis" option to submit your job. Within a few seconds, your task will be completed.

Viewing On-Screen Results
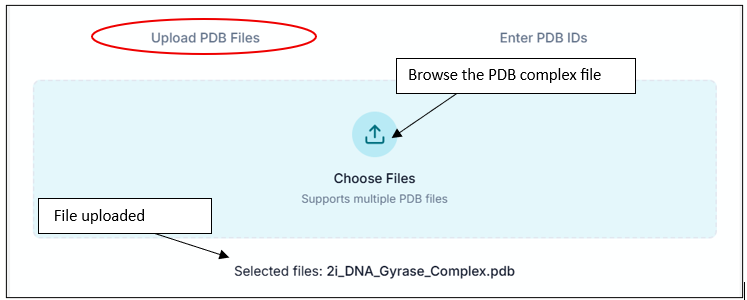
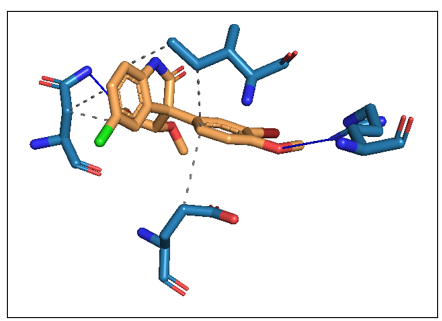
After successful completion, you can view the results in the "Results" section. This includes an interaction table, which contains all types of interactions your compound shows (e.g., hydrogen bonds, hydrophobic interactions). It also provides a 3D interaction image of the protein-ligand complex.
Downloading and Understanding the Output
You can download the results from the "Download Results" option from the same window. You will get a ZIP file that contains all your results.

Inside the ZIP folder, you will find several files, including:
- A protein-ligand complex PDB file.
- A folder containing intermediate files, including a PyMOL session file (
.pse) and a PNG image of the interaction. - A CSV file where all interactions are sorted and presented in a convenient tabular format.
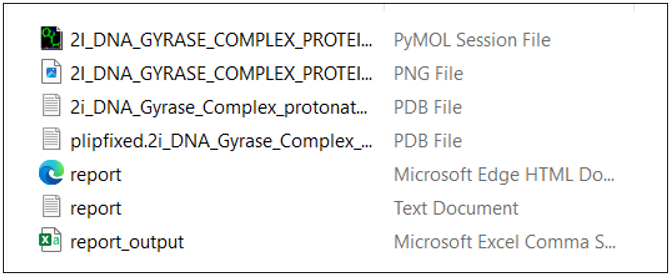
This is how the PyMOL session file looks. You can explore various options in PyMol, label the 3D interaction image, and use it as a publication-ready figure.
The report CSV file looks as shown in the image below.

