Introduction to SilicoCG
Conformer generation plays a vital role in drug discovery by capturing the three-dimensional flexibility of molecules, which is essential for accurately predicting how a drug interacts with its biological target. Since small molecules can adopt multiple low-energy conformations due to bond rotations, generating these conformers ensures that their most biologically relevant shapes are considered during virtual screening, docking, and molecular dynamics simulations.
This significantly improves the chances of identifying the correct binding pose and enhances hit rates in structure-based drug design. Conformer generation is also critical for pharmacophore modeling, as it allows better alignment of key functional groups across different molecules, leading to more accurate identification of potential drug candidates. Furthermore, the shape and flexibility of molecules influence their ADMET properties, making conformer generation valuable for predicting drug-like behavior. It also aids in scaffold hopping and lead optimization by helping researchers explore alternative chemotypes with similar 3D shapes, ultimately supporting the discovery of more effective and safer drug candidates.
A Step-by-Step Guide to Execute the Tool
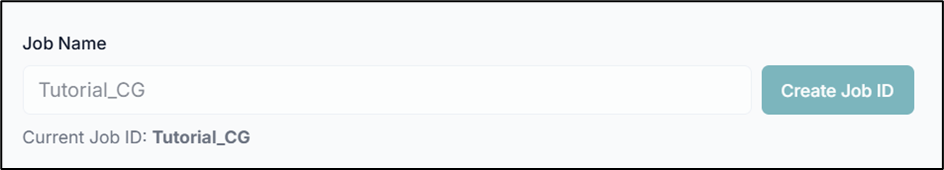
Before executing the tool, you must create a Job ID. You can customize this ID or click the "Create Job ID" button to have one generated for you automatically.
NOTE: Without creating a JOB ID, you will not be able to access any options of the tool.
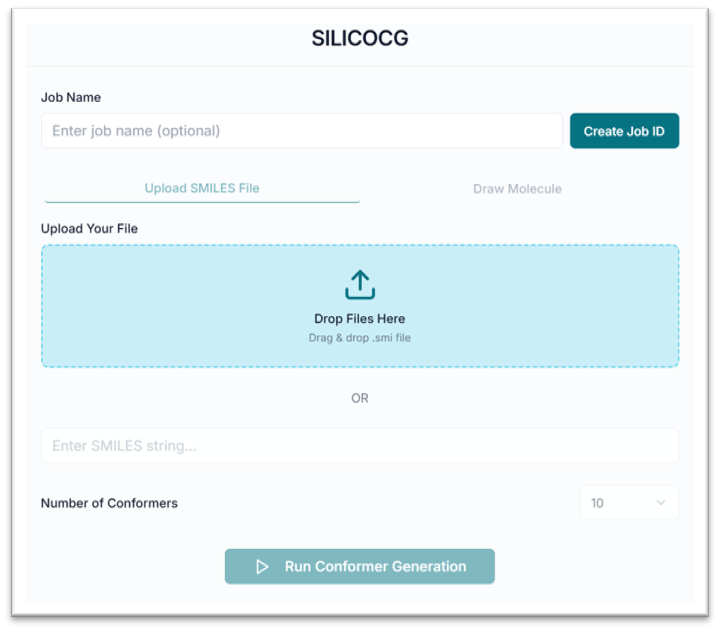
This is the SilicoCG tool's application workspace page, where various options for exploring and generating conformers are displayed. This tutorial will explain each option in detail.
Generating Conformers via SMILES (.smi) File
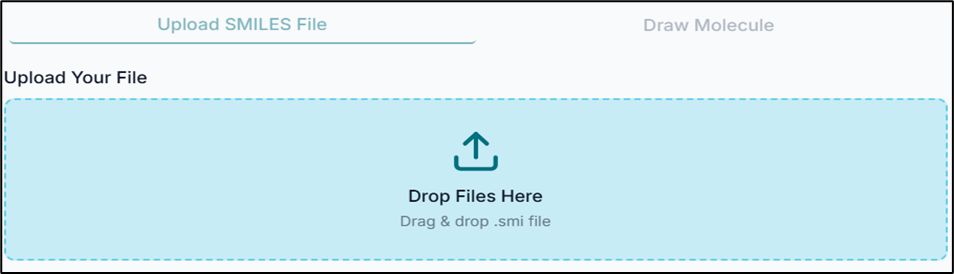
From the "Upload Your File" tab, click on the "Drop Files Here" option to upload your .smi file.
Browse through the files on your system and choose the SMILES file you want to upload. For example, here we use silicoCG_test.smi.
Your input .smi file should contain your compound SMILES and their respective names/IDs, in a format similar to the one shown below.

Tip: The SMILES string and the compound name should be TAB-separated.
Generating Conformers via Pasting SMILES
Alternatively, you can use the "SMILES Input" tab to paste your SMILES string directly into the text area.

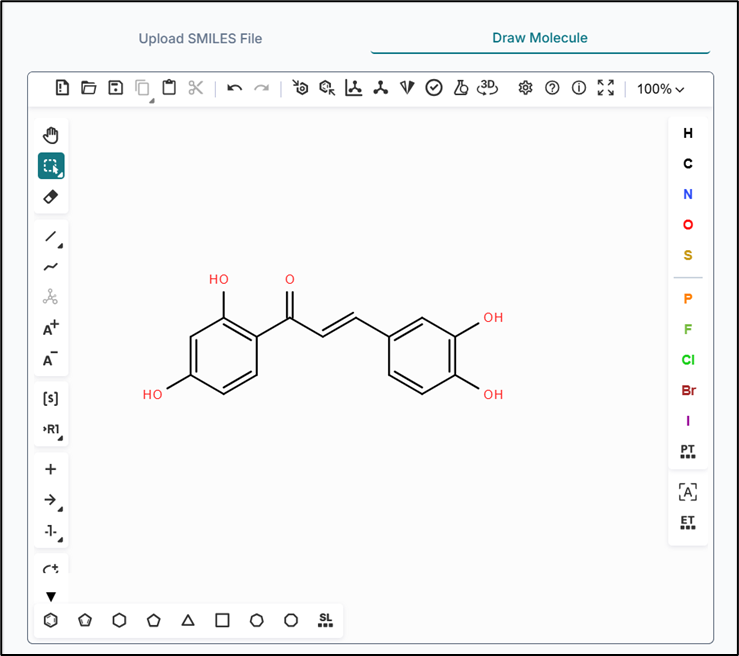
Generating Conformers via Drawing a Molecule
You can also draw any compound using the "Draw Molecule" option from the workspace window. The drawing tool provides multiple options to sketch your molecule as needed.

Configuring and Running the Job
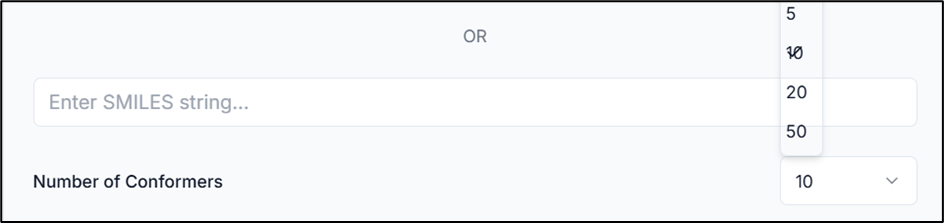
After providing your molecule, choose the number of conformers you want to generate by clicking on the "Number of Conformers" dropdown.

Tip: The minimum input for conformer generation is 5, and the maximum is 50.
To submit the job, click on the "Run Conformer Generation" button.
Downloading and Viewing Results
After your task completes successfully, a completion message will appear on the workspace.
You can download the results by clicking on the "Download Result" button.

A ZIP file containing the results will be downloaded. Each SDF file inside contains the respective conformers for each compound. For example, if you requested 10 conformers, the A1.sdf file will contain all 10 conformers for compound A1.
You can view all the generated conformers using our MolVisio tool. In the image below, all 10 conformers for compound A1 are visualized. Using MolVisio, you can also download the image, change its representation, and much more.

