Introduction to SilicoPeptify
SilicoPeptify is an advanced, AI-powered platform designed for the de novo design of novel bioactive peptides. It leverages a sophisticated transformer architecture to generate unique peptide sequences by combining three powerful approaches: designing based on desired physicochemical properties, integrating specific functional motifs, or creating peptides that are complementary to a protein target. This multifaceted approach empowers researchers to move beyond existing libraries, rapidly discover diverse therapeutic candidates, and accelerate the development of next-generation peptide-based drugs.
A Step-by-Step Guide to Execute the Tool
First, you must create a JOB ID before you can begin. You can enter a custom name for your job or simply click the "Create Job ID" button.
TIP: Access to all tool functions is disabled until a JOB ID has been successfully created.
Once created, your "New Job Created" confirmation will appear, along with your unique JOB ID. You are now ready to start designing peptides.
Method 1: Property-Based Generation
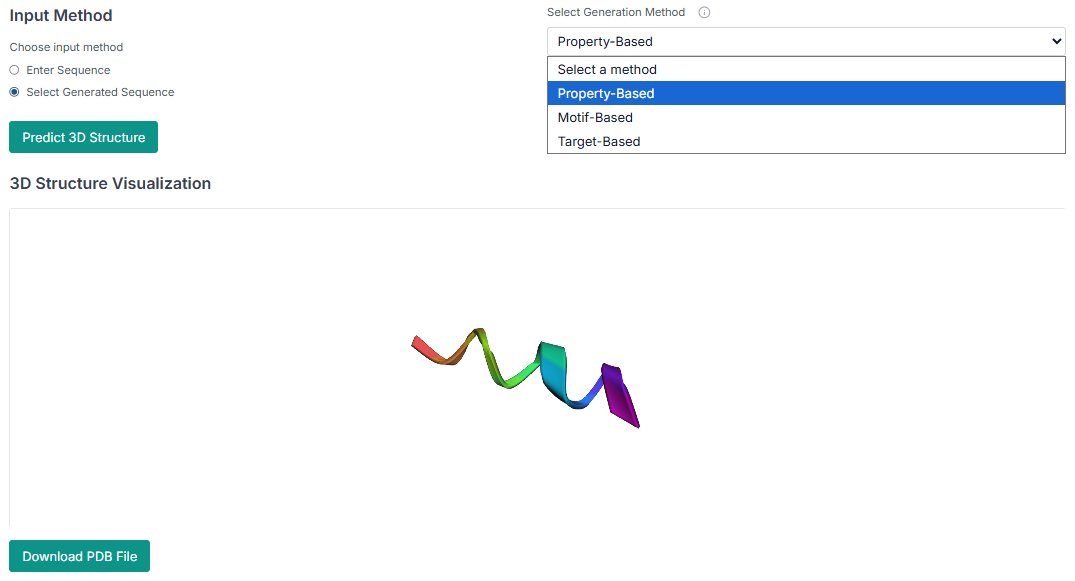
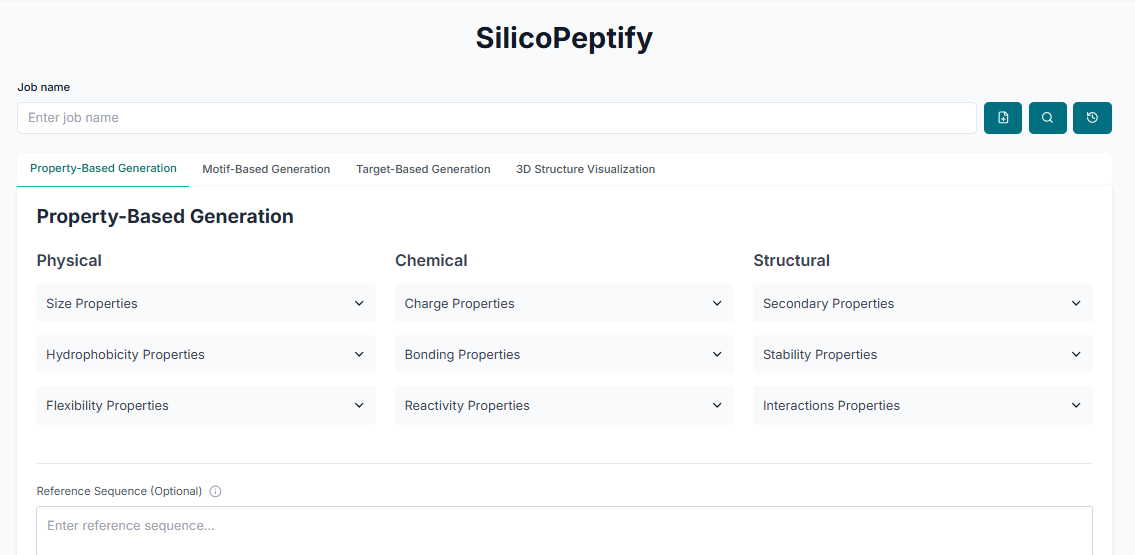
The first peptide generation method is Property-Based Generation. Select this option, and then choose the desired physicochemical properties (e.g., charge, hydrophobicity) that you want your generated peptides to have.
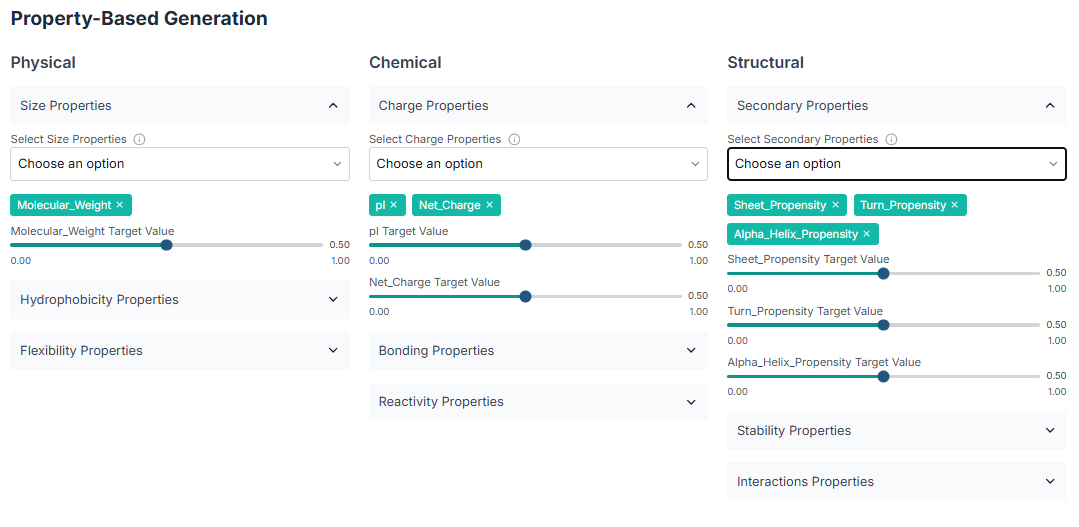
Next, configure the other parameters for the generation. Specify the desired "Sequence Length", the "Number of Peptides to Generate", and provide a reference sequence if you have one (this is optional).
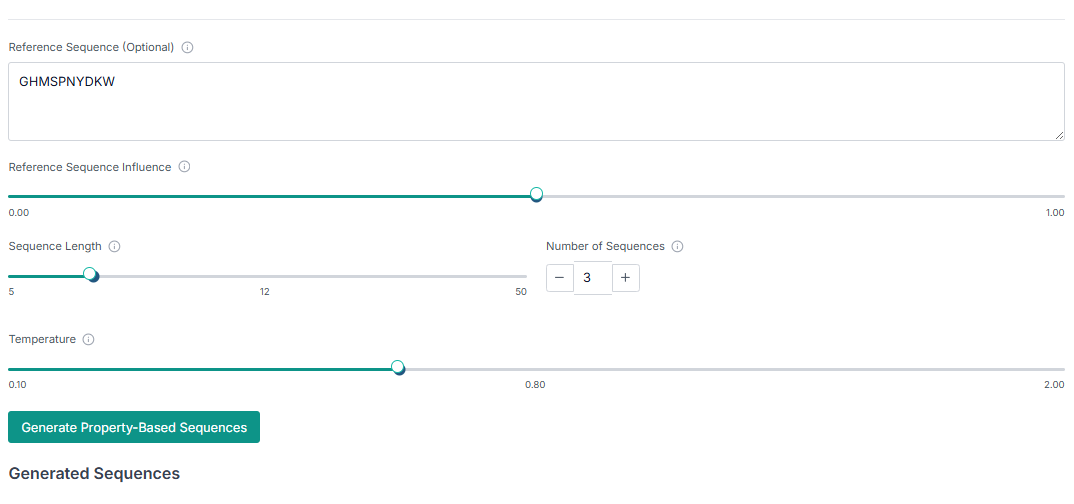
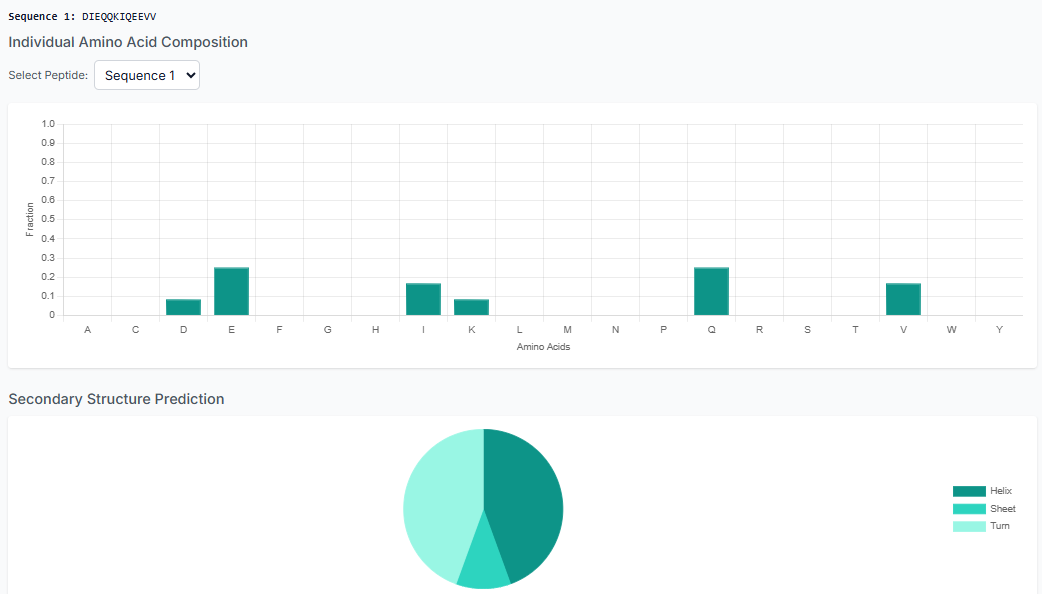
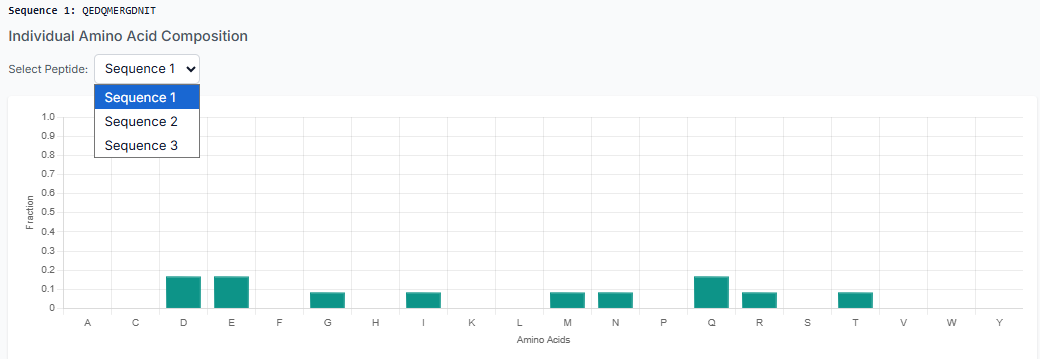
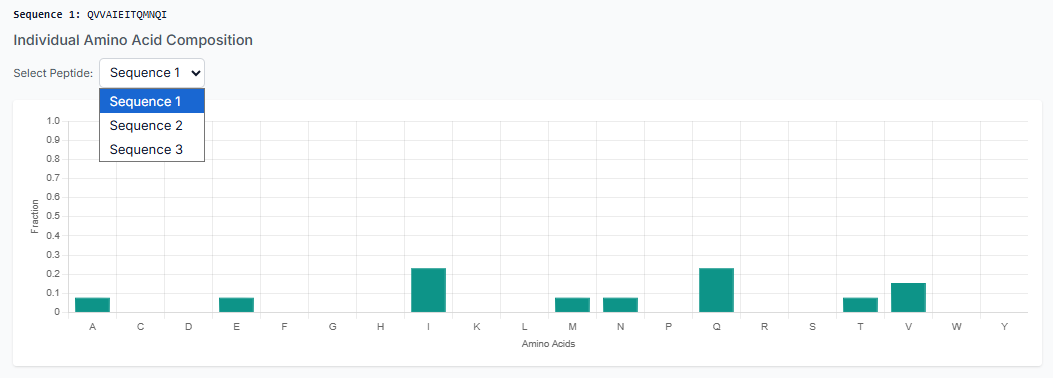
Once the peptides are generated, you can "Download the Generated Peptides" as a file. The results pane also allows you to visualize plots for composition and secondary structure analysis, giving you immediate insight into the generated library.
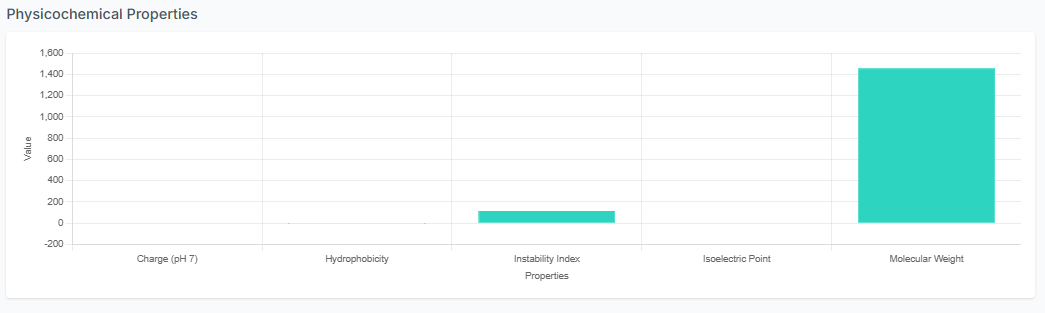
A separate plot is also visible for the physicochemical properties of the generated peptides. This allows for a comprehensive overview of your results and helps confirm that the peptides meet your design criteria.
Method 2: Motif-Based Peptide Generation
Choose this option to design peptides that are built around a specific, predefined functional motif.
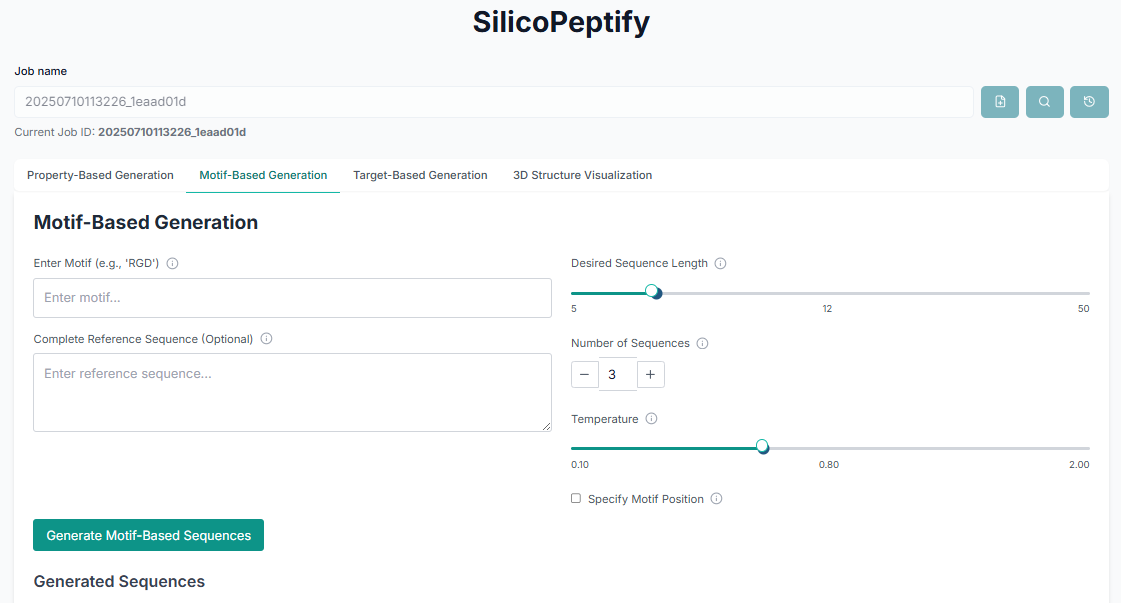
In this mode, you must "Provide the motif sequence" you wish to include. Then, configure its position within the final peptide and set other parameters like sequence length before generating the sequences.
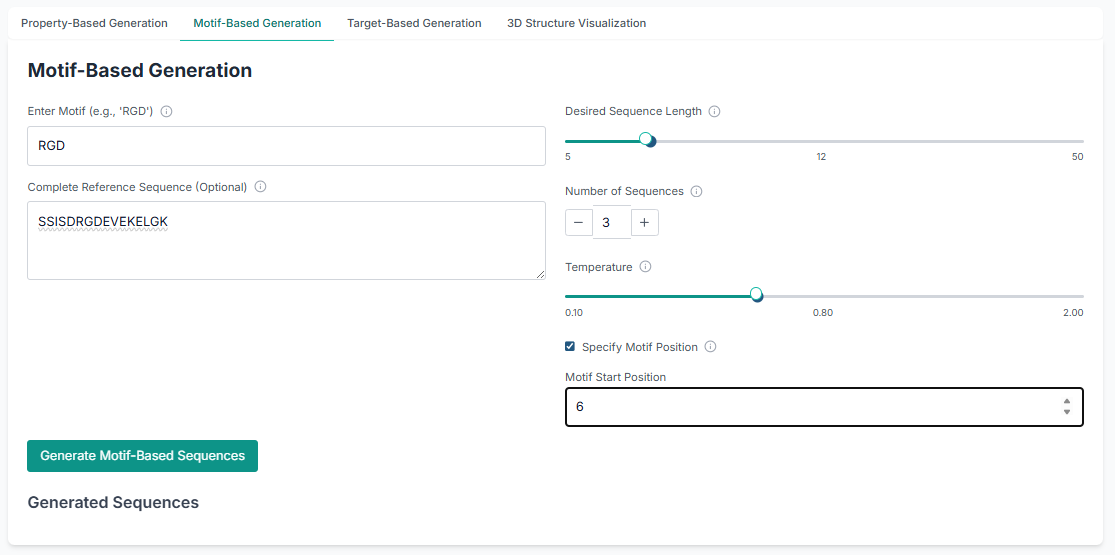
Similar to the first method, you can analyze and download the generated sequences. The results table and visualization plots will be available for your review and to confirm the successful integration of your motif.
Method 3: Target-Based Complementary Generation
Here, you provide the target sequence and specify the binding site positions for which a complementary peptide is to be generated. Then, click to generate the peptides.
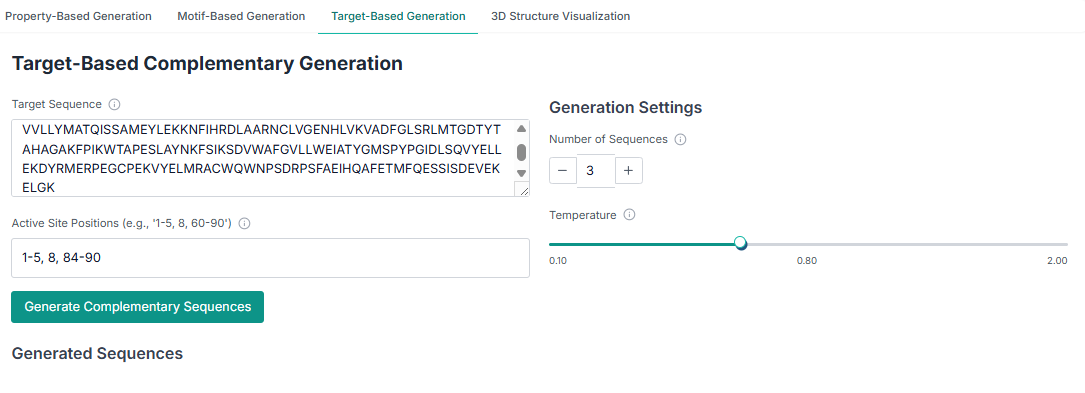
The same download and analysis capabilities are available for this method. You can save the generated peptide sequences and use the plots to analyze their properties.
3D Structure Prediction
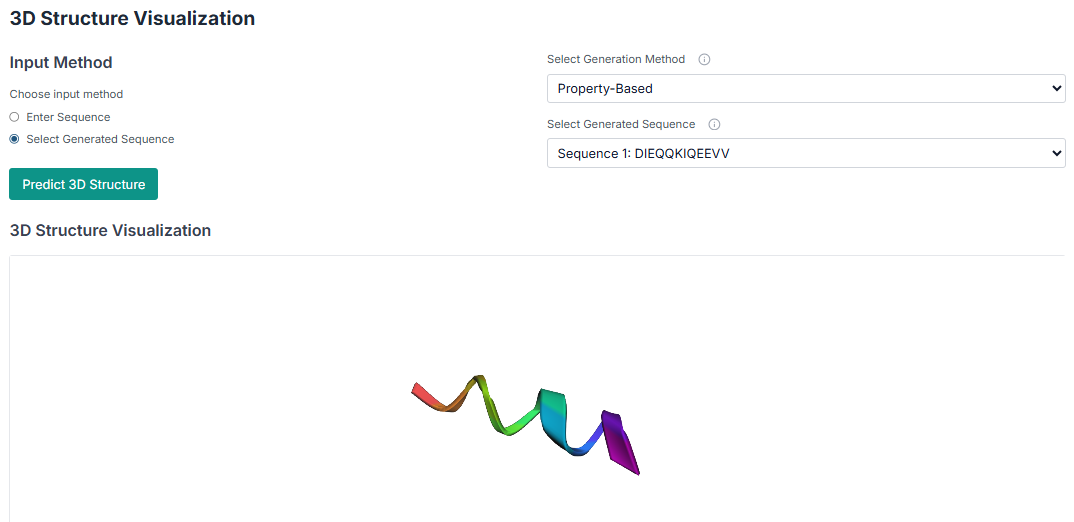
Beyond generation, SilicoPeptify can also predict the 3D structure of your peptides. You can either provide an input sequence manually or fetch a sequence from one of the generation jobs you just ran.
Select your peptide and predict its 3D structure, which can then be downloaded in PDB format.